News media write about genetics, genetic engineering and precision medicine in ways that might make us believe that our genomes are an open book. Genes and their functions appear well-annotated, and genome analysis has become routine for scientists.
In reality, the function of the vast majority of our DNA sequence is still poorly understood, if not unknown. Finding connections between genetic variation and disease or health will be a field of research for a long time to come.
To make matters worse, there is a bottleneck in the process of generating insight: genome analysis depends on the help of bioinformaticians, who are outnumbered by biologists at the ratio of at least eight to one.
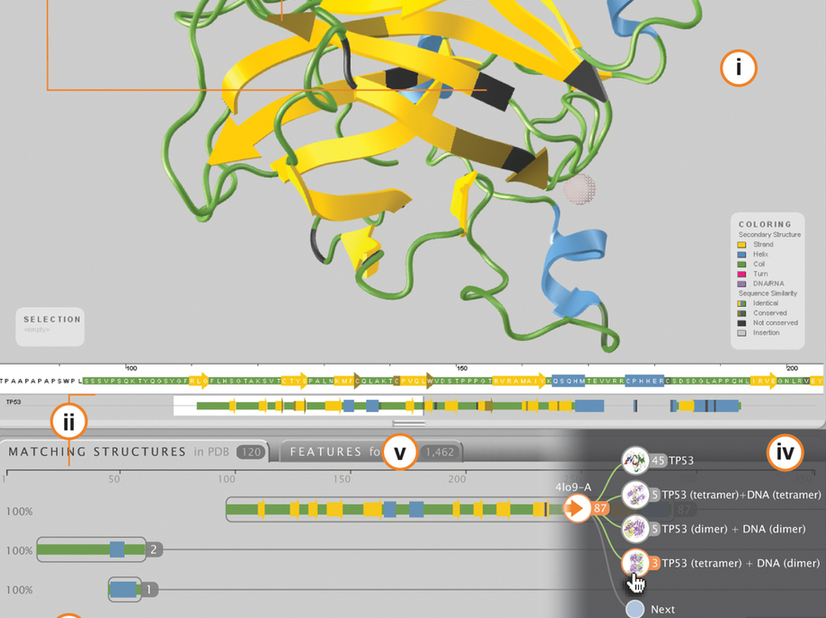
To make genomic data more accessible we built MetroNome, a web-based visual exploration platform. It presents genetic variation and gene expression data in an interactive context with information about the individuals’ health or disease and demographics.
Biologists can ask questions like “do mutations in gene X have an effect on the age of onset for disease Y?”. Without having to write code, scientists can use this platform to generate ideas and explore new hypotheses. MetroNome allows researchers to look at their own data and compare them with publicly available data sets, such as the 1,000 Genomes project or TCGA, the cancer gene atlas.